Question #649d2
2 Answers
Since the mRNA encodes for Proteins, the structure, i.e. the sequence of aminoacids would change. Depending on the specific mRNA sequence and the deleted base, you would obtain different proteins:
1.) if you delete a base it could give rise to a STOP-Codon (UAA, UGA, UAG) and your protein would be much shorter.
2.) the whole reading sequence gets shifted by one base and some base-triplets will be different than before, therefore your aminoacids will be different. This is called Frameshift.
Not sure what you mean by "Extra" base?
Explanation:
Anyway, mRNA is read in triplets (codons)..
Each triplet codes for a specific Amino Acid to be incorporated in the peptide chain. If one base would be deleted you would have what is called a "Frame Shift": starting with the triplet where the error occurs, the codons would be different and produce a different (and usually useless) protein....
Codon-table:
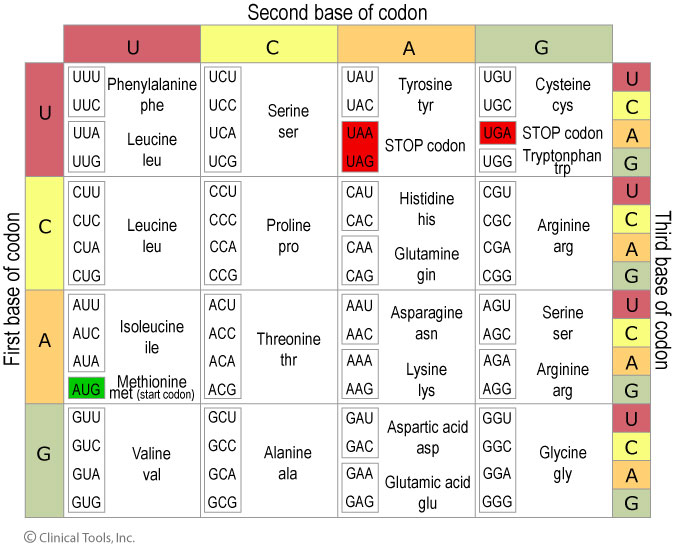 Copyright University of Leicester
Copyright University of Leicester
Example:
Original frame:
If in the original strand (as encoded in the DNA) somehow the red Cytosine is lost, the resulting frameshift will cause different AA's to be incorporated. In the example above it also results in a premature Stop-codon...


