How do you calculate molecular weight of DNA?
1 Answer
Here are the steps you might follow.
Explanation:
Step 1. DNA restriction
If you have a large DNA molecule, you will probably cut it into smaller fragments using a so-called DNA restriction enzyme.
Step 2. Gel electrophoresis
Then you will place properly prepared samples of the nucleic acid solution in the wells of an agarose gel electrophoresis system and apply a voltage for a specified amount of time.
In one of the wells, you also place a " DNA ladder": a sample that contains DNA fragments with known numbers of base pairs.
 DNA Ladder
DNA Ladder
(From www.slideshare.net)
The finished gel might look like this:
 archive.industry.gov.au
archive.industry.gov.au
The lanes on each end are the ladders. The distances travelled are proportional to the molar masses of the fragments.
Step 3. Calculate the numbers of base pairs in your fragments
Create a standard curve from your ladders by plotting log(base pairs) (bp) vs. distance travelled (d).
Assume your data were
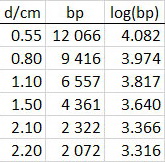 Data
Data
You would get a graph like this:
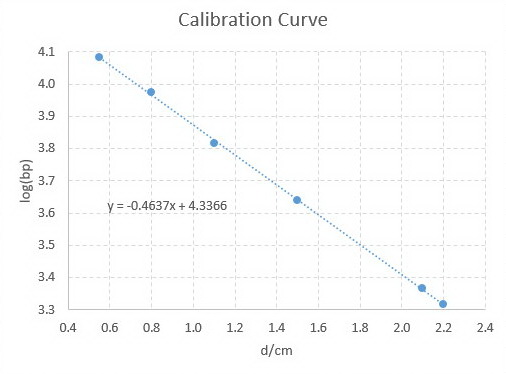 Calibration Curve
Calibration Curve
If one of your fragments had migrated 1.80 cm, you would have calculated
Step 4. Calculate the molar mass of the fragment.
The average mass of a base pair is 650 u.
Hence, the mass of the fragment is
and the molar mass is

